Liz Piotrowski scans for marine mammals in Western Passage using BigEyes.
This summer, undergraduate research assistants, Emma Newcomb and Liz Piotrowski, are leading a shore-based marine mammal visual monitoring program in Western Passage, a promising site for future tidal energy power. Once a week, they travel to Eastport, ME to conduct a 4-hour visual observation period, which later this summer will be supplemented by passive acoustic monitoring for marine mammal vocalizations (led by Chris Tremblay in Gayle Zydlewski’s lab). In their first month of observations, Emma and Liz have observed harbor porpoise, harbor seal, and minke whale.

Emma Newcomb looks out over Western Passage in search of marine mammals.
This research is conducted in collaboration with Ocean Renewal Power Company. Emma’s summer internship, which also includes a historical analysis of marine mammal stranding rates in Maine, is funded by the SEA Fellows program.

 Our proposal entitled, Assessing predator risk to diadromous fish conservation in the Penobscot River Estuary, has received funding from the
Our proposal entitled, Assessing predator risk to diadromous fish conservation in the Penobscot River Estuary, has received funding from the 

 Faythe’s analysis relied on long-term data collected by volunteers for the SC Department of Natural Resources, a program which Faythe herself has participated over the past 4 summers. Faythe is excited to spend one more summer on sea turtle patrol in SC before heading to the University of Georgia Veterinary School this fall.
Faythe’s analysis relied on long-term data collected by volunteers for the SC Department of Natural Resources, a program which Faythe herself has participated over the past 4 summers. Faythe is excited to spend one more summer on sea turtle patrol in SC before heading to the University of Georgia Veterinary School this fall. Faythe Goins presented her honors thesis research, Environmental Impacts on Loggerhead Sea Turtle Nesting on Edisto Island, SC, at the 2018 UMaine Student Symposium and won
Faythe Goins presented her honors thesis research, Environmental Impacts on Loggerhead Sea Turtle Nesting on Edisto Island, SC, at the 2018 UMaine Student Symposium and won 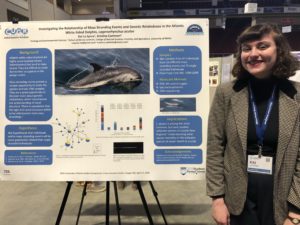 Kai LaSpina presented preliminary results from her Capstone research, Investigating the relationship of mass stranding events and genetic relatedness in the Atlantic white-sided dolphin, Lagenorhynchus acutus, at the
Kai LaSpina presented preliminary results from her Capstone research, Investigating the relationship of mass stranding events and genetic relatedness in the Atlantic white-sided dolphin, Lagenorhynchus acutus, at the  This past month has been a wonderful whirlwind of work-related travel, to and from Canada (twice!) and roundabout Maine. I kicked off the month with a trip to the University of New Brunswick in St. John to give an invited seminar to the Biology Department. The following week, I ferried across the Bay of Fundy to the Society for Marine Mammalogy biennial conference held this year in Halifax. Finally, earlier this week I gave a seminar at Maine Maritime Academy.
This past month has been a wonderful whirlwind of work-related travel, to and from Canada (twice!) and roundabout Maine. I kicked off the month with a trip to the University of New Brunswick in St. John to give an invited seminar to the Biology Department. The following week, I ferried across the Bay of Fundy to the Society for Marine Mammalogy biennial conference held this year in Halifax. Finally, earlier this week I gave a seminar at Maine Maritime Academy.