Happy New Year! Interested in learning about some of the scientific discoveries related to marine mammals from the past year. Check out the annual Marine Mammals in the News online publication, produced by undergraduate students in INT308 (Marine Mammal Ecology and Conservation). From whales to manatees, from the Arctic to the Antarctic, from conservation to physiology, and much more – check out the stories they wrote, highlighting marine mammal science that has been published within the last year.
Season’s greetings and Happy New Year from the Cammen lab! Check out UMaine’s season’s greetings video, featuring our very own Alayna Hawkins (at 0:42 into the video), in a photo taken by our very own Holland Haverkamp. It’s great to see our students representing what lab work can look like at the University of Maine.
Best wishes to all for a happy, healthy, and productive 2019!
Check out the latest issues of NOAA Fisheries Science Connect, which features our recent publication in Ecology and Evolution. Science Connect highlights the latest publications focused on protected species science.
Congratulations to Emma Newcomb and Liz Piotrowski on receiving fellowships from the UMaine Center for Undergraduate Research (CUGR)! Both Emma and Liz were named AY 2018-19 CUGR Fellows. Their fellowships will support research in: “Effects of human interaction on marine mammal strandings and call reports in the Gulf of Maine from 2010 to 2015” (Emma) and “Using eDNA sampling as a mechanism for improving marine mammal conservation through a non-invasive and cost efficient technique” (Liz).
As part of a new, collaborative NSF-funded research and training program in the genomic ecology of coastal organisms, we are recruting a PhD student who will study genome-phenome relationships in the wild. The student will be expected to conduct both field work and genomic analyses towards understanding adaptation and the links between plumage phenotypes (color, resilience to wear, and microbiomes) and reproductive fitness across sparrow species. Field work during summer months may involve supervising field crews in tidal marshes across the Northeast US, from Maine to Virginia. Genomic analyses will include candidate gene sequencing, gene expression analyses, and microbiome characterization.
The student will be co-advised by Drs. Kristina Cammen (http://cammenlab.org) and Brian Olsen (https://sbe.umaine.edu/olsen-2/), through the Ecology and Environmental Sciences program at the University of Maine, located in Orono, an hour to the ocean and an hour and a half to Maine’s highest peak. The student will also have the opportunity to work in collaboration with a diverse team of investigators, graduate students, and undergraduate students at the Universities of New Hampshire and Maine studying the ecological genomics and eco-evolutionary feedbacks of adaptation in tidal marsh birds.
The successful candidate must have a strong background in ecology and/or genomics. Preferred candidates will have demonstrated experience with field work, in particular, bird mist-netting (previous time as a federal banding sub-permittee strongly preferred), as well as experience in genetics, genomics, and/or bioinformatics. Consistent with our program scope and to advance an integrated understanding of adaptation in nature, we are especially interested in candidates who show promise to work in an inclusive and diverse collaborative environment and to engage intellectually across the diverse scales of genomes, phenomes, and environmental feedbacks. Individuals who are intellectually curious, responsible, willing to learn, team-oriented, and have attention to detail are encouraged to apply. An M.S. in a related field is preferred, but qualified candidates with extensive experience will be considered.
To apply, please send a cover letter describing your qualifications, including your commitment to diversity and inclusion in collaborative science, a curriculum vitae, unofficial transcripts, and the contact information for at least three references to kristina.cammen@maine.edu and brian.olsen@maine.edu with “Ecological Genomics PhD Student Search” as the subject line of your email. All applications received before November 14, 2018 will receive full consideration, and applications will be accepted on a rolling basis thereafter until the positions are filled. A start date of January 2019 is strongly preferred.

(left to right) Lynda Doughty, Alayna Hawkins, Sarah Burton, Shannon Brown, Emma Newcomb, and Emma Spies at the Marine Mammals of Maine triage facility in Harpswell, ME.
Lynda Doughty, executive director and stranding coordinator, of Marine Mammals of Maine hosted a visit of several of our lab members to MMoME’s seal triage and rehabilitation center this past weekend. This was a particularly exciting visit for our students who have hours and hours of experience working with marine mammal stranding data in our custom-built electronic database, and our students who spend most of their time at a lab bench working with seal DNA. Lynda gave us a tour of the facility and shared stories and lessons learned from the past several months of crisis response to a federally-designated Unusual Mortality Event that more than tripled the number of seals they were called to respond to, compared to previous years. We talked about several ways to strengthen our partnership between the University of Maine and MMoME, with a particular focus on how students can be involved. To support one ongoing research collaboration, we also collected a water sample from a seal rehab pool for continued eDNA (environmental DNA) method development. Thank you, Lynda, for hosting our visit!

Sarah Burton collects a water sample for eDNA analysis.
Emma Newcomb, a 2018 SEA Fellow and Cammen Lab undergraduate research assistant, presented her research at a public, student-focused symposium held at the Darling Marine Center in Walpole, Maine earlier this week. The symposium was attended by >80 scientists, industry and community members, and the interested general public. Over 20 undergraduate students presented posters about their summer research experience, all linked by a shared goal to improve our understanding of the Maine’s coastal ecosystem to help benefit our coastal communities and economies. You can read more about the 2018 SEA Fellows Symposium here.
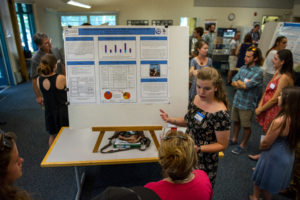
Photo by Holland Haverkamp
Emma’s research focused on classifying and quantifying marine mammal-human interaction cases that are observed in Maine and recorded by our collaborating stranding organizations, Marine Mammals of Maine and Allied Whale. Human interaction cases are currently categorized nationally as boat collision, gunshot, fisheries interaction, and “other”, and we are working to further describe this hard-to-define “other” category, which is this the most common type of human interaction for our stranded marine mammals in Maine. These interactions are normally typified by a human approach within 150 yards and oftentimes referred to as marine mammal harassment.
We would like to acknowledge the University of Maine System’s Research Reinvestment Fund (RRF) for their support of Emma’s summer research experience, part of an Interdisciplinary Undergraduate Research Collaborative grant. Emma will continue her research in this area during the academic year, supported by a NOAA Prescott grant.
This past week, Dr. Kristina Cammen and undergraduate research assistant, Liz Piotrowski, collected their first eDNA water samples at the Marine Mammals of Maine (MMoME) triage facility located in Harpswell, Maine. Thank you to Dominique Walk, MMoME’s assistant stranding coordinator, and several MMoME interns for their collaboration in this effort.
eDNA, short for environmental DNA, is free floating DNA that is left behind by an organism as it travels through an environment. We are working to develop protocols that will allow us to isolate and characterize this DNA, for example from a water sample collected in a seal rehab pool and ultimately from seawater samples collected near seal haul-out sites. Our hope is that these non-invasive sampling methods will enable future investigations of the genetic diversity and population structure of seals in the Gulf of Maine.
9/29/18: Updated to add that our first attempts to detect and sequence seal DNA from the MMoME facility have been successful! We are now working to optimize our protocols so that we can distinguish between individuals that are present in the same pool of water.
The Cammen Lab welcomes their newest member, Alayna Hawkins, who begins an MS in Marine Biology program this summer. A recent graduate of St. Francis College in Pennsylvania, with an interest in marine mammal science and genetics, Alayna joins the lab to conduct research on gray and harbor seal immunogenetics. She hopes to help elucidate why gray seals appear more resistant than harbor seals to several viral diseases through an exploration of the genetic diversity of their immune systems.
The recent recovery of gray and harbor seals in the Northwest Atlantic, following historical exploitation and subsequent protection, provides a natural “experiment” in which to evaluate the impact of changes in population size and distribution on genetic diversity. It is a rare opportunity to test evolutionary theories, for example that genetic bottlenecks will reduce diversity (in extreme cases, leading to inbred populations), in a natural population. With a genomic approach, called RAD sequencing, we evaluated how diversity has changed over time and space in multiple cohorts of gray and harbor seals sampled over the past half-century. Our findings clearly show that signatures of historical bottleneck remain in the genomes of the species today, but also find high contemporary diversity, suggesting the species are not inbred. Interestingly, we find higher diversity in gray seals than harbor seals, which may have important implications for species fitness, a point we’d like to continue to investigate moving forward.
Co-authors on this paper include collaborators from the Duke University Marine Lab, NOAA NEFSC, Canada DFO, Tufts University, and St. Mary’s University. If you’re interested in other research we’ve done on this topic, check out our comparison of contemporary and archaeological seals from this region.
A full, freely available copy of our new open-source paper can be found here, or feel free to contact me directly for more information.
Cammen KM, Bowen WD, Hammill MO, Puryear WB, Runstadler J, Wenzel FW, Wood SA, Frasier TR, Kinnison M (accepted) Genomic signatures of population bottleneck, recovery, and expansion in Northwest Atlantic pinnipeds. Ecology and Evolution.